As mentioned in a previous post, we attended the HUPO 11th Annual World Congress in Boston recently. I was lucky enough to attend so I can share some of the highlights from each day with you.
The HUPO council meeting was held on Sunday. Here HUPO shared its vision for the future. One aim is to expand the involvement of researchers at all levels and across different disciplines. This was reflected in the main scientific program for the rest of the week, which included talks on metabolomics, lipidomics and analysis of non-human samples.
The New Technology and Standardisation in Proteomics workshop was also held on Sunday, covering varied topics, with each talk showcasing different aspects of experimental procedure: from sample preparation to data analysis. Due to a last minute change, I was drafted in to chair the afternoon session of the workshop. As part of this, I got to introduce Roy Martin from Waters with his presentation on the TransOmics™ Informatics software for proteomics and metabolomics that we have covered in an earlier blog post.
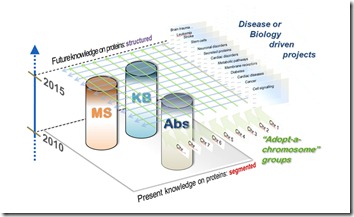
Launching the Human Proteome Project
On Monday, a major event was the official launch of the chromosome-based Human Proteome Project (C-HPP). This 10 year multi-site research effort was celebrated with a well-attended session that introduced the potential and challenges of this project. There were informal presentations from the major groups involved, including the leadership team, principal investigators from the labs doing the wet work, funding organisations such as the NIH, journals and the HUPO Industrial Advisory Board, of which Nonlinear are members. It was good to be there to hear what was expected and celebrate with the cutting of the HUPO cHPP cake!
 Principle investigators, leadership group and other organisations involved cut the cake to launch the C-HPP
Principle investigators, leadership group and other organisations involved cut the cake to launch the C-HPP
The goal of the C-HPP is to produce what’s been called a “parts list”. Each lab in the project has taken on the task of characterising the proteins that map to the genes from a specific chromosome within the human body. Supporting this are technology pillars that add context to these discoveries. There were words of praise and encouragement, but also a reality check on how industry needs to help drive this forward, much like the efforts of the Human Genome Project, and how the C-HPP must not suffer the same pitfalls as other big projects. The meeting ended on a positive note – everyone should have access to the data and it can be used by and built upon by the next generation of proteomics researchers.
Posters and Progenesis interest
 On Tuesday I presented a poster (#108) showing how you can use gas-phase fractionation to increase proteome coverage and how Progenesis LC-MS supports that. If you’re interested to learn more about this too, then a good start is this blog post.
On Tuesday I presented a poster (#108) showing how you can use gas-phase fractionation to increase proteome coverage and how Progenesis LC-MS supports that. If you’re interested to learn more about this too, then a good start is this blog post.
There were over 600 other posters shown at the meeting, with an equal number of label-free quantification compared to labelled approaches for LC-MS. 2D gels were also represented, mainly as a way of separating out proteins prior to complementary analysis by MS technologies.
Progenesis LC-MS and Progenesis SameSpots were specifically mentioned on 9 and 2 posters respectively but we also had interest in our newest product, Progenesis CoMet, for metabolomics. If you would like to try this new product to see how it can help complement your research, then please get in touch and we’ll be happy to help.
Find out more about Progenesis LC-MS
As you can see in the photo above, the exhibit hall was busy each day, especially on Tuesday and Wednesday evenings. By the final day we had answered a lot of questions from our customers about exporting and reimporting results to and from other bioinformatics packages. This is an area I feel we are quite good at, so it was good to share what we knew and if you have any questions, please get in touch and our support team can help you.
We also had quite a few enquiries from people working on quantifying proteins with post-translational modifications and separately analysis of native peptides. Progenesis LC-MS is being used for both of these applications by our current customers. If you would like details, then contact us for a demo on your data.
Next year the meeting will in Yokohama, Japan, with the tentative theme so far of “The Evolution of Technology in Proteomics”, which sounds like an interesting meeting for us. We hope to see you there to share our latest technologies for ‘omics data analysis.
View from the 50th Floor of the Prudential Building, looking over Boston at the HUPO 2012 evening social